Lab 7
MC Questions
Practical Questions
- Question 4
- A k-fold cross-validation can be used to determine the best choice of hyper-parameters from a finite set of choices – True
- Question 8
- Maximum depth, minimum node size, and learning rate are all examples of what type of parameters? – Hyper-parameters
- Question 9
- Selecting an appropriate model requires – Both knowing the nature of your problem and the nature of your model (in terms of linearity).
- Question 10
- A good reason for implementing a feature selection technique is – All of the above
- Question 11
- The concept of principal component analysis refers to – Determining the directions along which we maximize the variance of the input features.
Preprocessing: Diabetes Dataset
columns = 'age gender bmi map tc ldl hdl tch ltg glu'.split()
diabetes = datasets.load_diabetes()
df = pd.DataFrame(diabetes.data, columns=columns)
y = diabetes.target
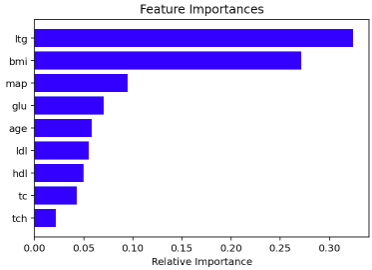
Question 1
rf_class = RandomForestRegressor(random_state=310, max_depth=10, n_estimators = 1000)
rf_class.fit(df,y)
features = df.columns
importances = rf_class.feature_importances_
indices = np.argsort(importances)[-9:]
plt.title('Feature Importances')
plt.barh(range(len(indices)), importances[indices], color='b', align='center')
plt.yticks(range(len(indices)), [features[i] for i in indices])
plt.xlabel('Relative Importance')
plt.show()
Question 2
def stepwise_selection(X, y,
initial_list=[],
threshold_in=0.01,
threshold_out = 0.05,
verbose=True):
""" Perform a forward-backward feature selection
based on p-value from statsmodels.api.OLS
Arguments:
X - pandas.DataFrame with candidate features
y - list-like with the target
initial_list - list of features to start with (column names of X)
threshold_in - include a feature if its p-value < threshold_in
threshold_out - exclude a feature if its p-value > threshold_out
verbose - whether to print the sequence of inclusions and exclusions
Returns: list of selected features
Always set threshold_in < threshold_out to avoid infinite looping.
See https://en.wikipedia.org/wiki/Stepwise_regression for the details """
included = list(initial_list)
while True:
changed=False
# forward step
excluded = list(set(X.columns)-set(included))
new_pval = pd.Series(index=excluded)
for new_column in excluded:
model = sm.OLS(y, sm.add_constant(pd.DataFrame(X[included+[new_column]]))).fit()
new_pval[new_column] = model.pvalues[new_column]
best_pval = new_pval.min()
if best_pval < threshold_in:
best_feature = new_pval.idxmin()
included.append(best_feature)
changed=True
if verbose:
print('Add {:30} with p-value {:.6}'.format(best_feature, best_pval))
# backward step
model = sm.OLS(y, sm.add_constant(pd.DataFrame(X[included]))).fit()
# use all coefs except intercept
pvalues = model.pvalues.iloc[1:]
worst_pval = pvalues.max() # null if pvalues is empty
if worst_pval > threshold_out:
changed=True
worst_feature = pvalues.idxmax()
included.remove(worst_feature)
if verbose:
print('Drop {:30} with p-value {:.6}'.format(worst_feature, worst_pval))
if not changed:
break
return included
result = stepwise_selection(X_d,y_d,[],0.001,0.001)
Question 3
Xs_d = scale.fit_transform(X_d)
model = ElasticNet(alpha=0.1, l1_ratio=0.5)
model.fit(Xs_d, y_d)
model.coef_
list(X_d.columns[np.abs(model.coef_)>1e-1])
X_d.shape[1]
v = -np.sort(-np.abs(model.coef_))
for i in range(X_d.shape[1]):
print(X_d.columns[np.abs(model.coef_)==v[i]])
Preprocessing Breast Cancer Dataset
data = load_breast_cancer()
df = pd.DataFrame(data.data, columns=data.feature_names)
feats = ['mean radius', 'mean texture']
X = df[feats].values
y = data.target
Question 5
model = tree.DecisionTreeClassifier(random_state=1693)
params = [{'max_depth':np.linspace(1, 100, 100).astype(int),
'min_samples_leaf':np.linspace(1, 25, 25).astype(int)}]
gs = GridSearchCV(estimator=model,cv=10,scoring='accuracy',param_grid=params)
gs_results = gs.fit(X, y)
print(gs_results.best_params_) # --> {'max_depth': 4, 'min_samples_leaf': 23}
print(f'Accuracy is: {gs_results.best_score_}')
Question 6
# do k fold and parameter filtering
model = tree.DecisionTreeClassifier(random_state=12345)
params = [{'max_depth':np.linspace(1, 100, 100).astype(int),
'min_samples_leaf':np.linspace(1, 25, 25).astype(int)}]
gs = GridSearchCV(estimator=model,cv=10,scoring='neg_mean_squared_error',param_grid=params)
gs_results = gs.fit(X, y)
print(gs_results.best_params_)
# output false negatives
dt_class = tree.DecisionTreeClassifier(random_state=12345, max_depth=4, min_samples_leaf=23)
dt_class.fit(X, y)
y_b_pred = dt_class.predict(X)
dt_cm = confusion_matrix(y, y_b_pred)
dt_cm
Question 7
# do k fold and parameter filtering
model = tree.DecisionTreeClassifier(random_state=1693)
params = [{'max_depth':np.linspace(1, 100, 100).astype(int),
'min_samples_leaf':np.linspace(1, 25, 25).astype(int)}]
gs = GridSearchCV(estimator=model,cv=10,scoring='neg_mean_squared_error',param_grid=params)
gs_results = gs.fit(X, y)
print(gs_results.best_params_)
# output accuracy
def validation(X,y,k,model):
PA_IV = []
PA_EV = []
pipe = Pipeline([('scale',scale),('Classifier',model)])
kf = KFold(n_splits=k,shuffle=True,random_state=1693)
for idxtrain, idxtest in kf.split(X):
X_train = X[idxtrain,:]
y_train = y[idxtrain]
X_test = X[idxtest,:]
y_test = y[idxtest]
pipe.fit(X_train,y_train)
PA_IV.append(accuracy_score(y_train,pipe.predict(X_train)))
PA_EV.append(accuracy_score(y_test,pipe.predict(X_test)))
return np.mean(PA_IV), np.mean(PA_EV)
model = tree.DecisionTreeClassifier(random_state=1693, max_depth=4, min_samples_leaf=23)
validation(X, y, 10, model) # --> 89%
Question 12
df12 = pd.DataFrame(data.data, columns = data.feature_names)
X = df12.values
y = data.target
pca = PCA(n_components=9)
model = tree.DecisionTreeClassifier(random_state=1234, max_depth=10, min_samples_leaf=20)
principalComponents = pca.fit_transform(X)
model.fit(principalComponents, y)
y_pred = model.predict(principalComponents)
dt_cm = confusion_matrix(y, y_pred)
dt_cm